The Browne Lab
UNC HIV Cure Center
Genetic Medicine Research Building
120 Mason Farm Road
Chapel Hill, NC 27599
Accepting Students
Postdoc Position Available
Research Summary
We study the molecular mechanisms of HIV latency. Transcriptional silencing of HIV is a key mechanism of persistence in patients, and is a barrier to viral eradication, but little is known about the latent reservoir or the molecular mechanisms that regulate it. As such, our repertoire of drugs for targeting latently infected cells is limited. Some latency reversing agents (LRAs) have been developed, but these are typically reactivate only a minor subset of proviruses. This inefficiency is in part due to the reservoir not constituting a uniform target, but instead being a heterogeneous set of cells with diverse characteristics and restrictions to HIV expression. However, most analyses of latency use bulk cell cultures assays in which crucial information about the behavior of individual cells is lost. Also, latently infected cells in patient samples are exceedingly rare, making them very difficult to study directly. New technological breakthroughs in the field of single cell analysis as well as the development of primary cell models for HIV latency now open the possibility of observing how latently infected cells form and are maintained at single cell resolution. Our lab has developed tools to study the establishment, maintenance and reversal of HIV latency at single cell resolution using multi-omics methods. Furthermore, we combine these approaches with genetic perturbation, time-lapse microscopy and novel bioengineering tools to gain insight into how the host cell regulates HIV latency. We have recently discovered using single cell RNAseq (scRNAseq) that latency in primary CD4 T cells is associated with expression of a distinct transcriptional signature (Bradley et al 2018). Our hypothesis is that this signature represents part of a cellular program that regulates latency, and that this program is an exciting novel target for the development of LRAs. Ongoing projects in the lab involve the application of new technologies to our model systems, and testing/validation of the roles of host cell pathways we have identified in HIV latency. Our overall goal is to identify new targets for the development of drugs to clear the HIV reservoir.
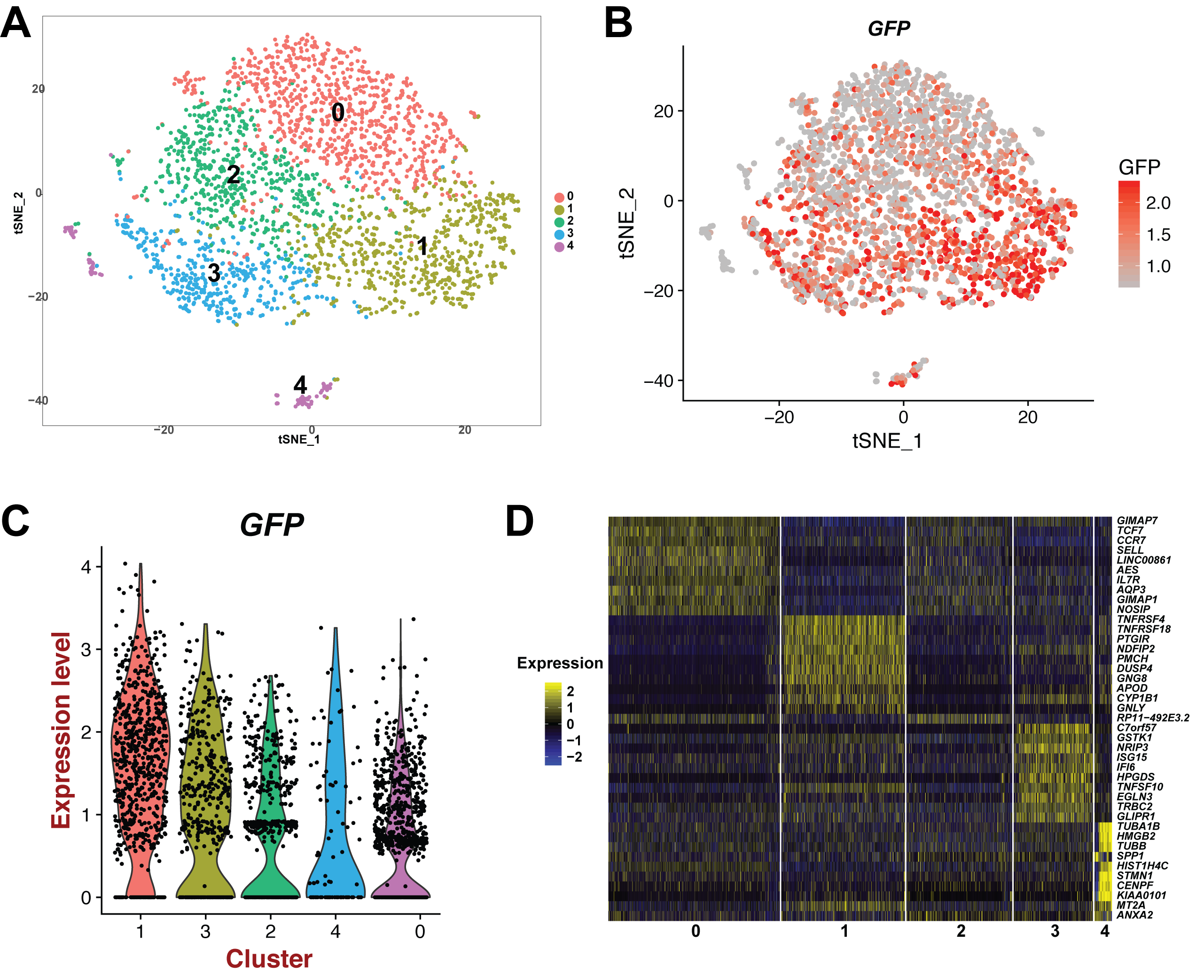
Single cell RNAseq of HIV infected CD4 T cells
Selected Publications
(click here for a full publication list)
Reyes M, Vickers D, Billman K, Eisenhaure T, Hoover P, Browne EP, Rao DA, Hacohen N, Blainey PC. (2019) Science Advances. Jan 23;5(1) Multiplexed enrichment and genomic profiling of peripheral blood cells reveal subset-specific immune signatures.
Mahoney, SA. Shukla, N Patsoukis, A Chaudhri, EP. Browne, A Arazi, T Eisenhaure, WF. Pendergraft III, P Hua Hung C. Pham, X Bu, B Zhu, N Hacohen, E Fritsch, VA Boussiotis, CJ Wu, G Freeman. (2018 Cancer Cell -In press) A secreted PD-L1 splice variant that covalently dimerizes and mediates immunosuppression
Bradley T, Ferrari G, Haynes BF, Margolis DM, Browne EP. Cell Rep. 2018 Single-Cell Analysis of Quiescent HIV Infection Reveals Host Transcriptional Profiles that Regulate Proviral Latency. Oct 2;25(1):107-117.e3. doi: 10.1016/j.celrep.2018.09.020.
Donlin LT, Rao DA, Wei K, Slowikowski K, McGeachy MJ, Turner JD, Meednu N, Mizoguchi F, Gutierrez-Arcelus M, Lieb DJ, Keegan J, Muskat K, Hillman J, Rozo C, Ricker E, Eisenhaure TM, Li S, Browne EP, Chicoine A, Sutherby D, Noma A; Accelerating Medicines Partnership RA/SLE Network, Nusbaum C, Kelly S, Pernis AB, Ivashkiv LB, Goodman SM, Robinson WH, Utz PJ, Lederer JA, Gravallese EM, Boyce BF, Hacohen N, Pitzalis C, Gregersen PK, Firestein GS, Raychaudhuri S, Moreland LW, Holers VM, Bykerk VP, Filer A, Boyle DL, Brenner MB, Anolik JH. (2018). Methods for high-dimensonal analysis of cells dissociated from cyropreserved synovial tissue. Arthritis Res Ther. 2018 Jul 11;20(1):139. doi: 10.1186/s13075-018-1631-y.
Letham B, Letham PA, Rudin C, Browne EP (2016). Prediction uncertainty and optimal experimental design for learning dynamical systems. Chaos. 2016 Jun;26(6):063110. doi: 10.1063/1.4953795.
Cadena C, Stavrou S, Manzoni T, Iyer SS, Bibollet-Ruche F, Zhang W, Hahn BH, Browne EP, Ross SR (2016). The effect of HIV-1 Vif polymorphisms on A3G anti-viral activity in an in vivo mouse model. Retrovirology. 2016 Jun 30;13(1):45. doi: 10.1186/s12977-016-0280-y.
Browne EP, Letham B, Rudin C (2016). A Computational Model of Inhibition of HIV-1 by Interferon-Alpha. PLoS One. 2016 Mar 24;11(3):e0152316. doi: 10.1371/journal.pone.0152316. eCollection 2016.
Tas JM, Mesin L, Pasqual G, Targ S, Jacobsen JT, Mano YM, Chen CS, Weill JC, Reynaud CA, Browne EP, Meyer-Hermann M, Victora GD (2016) . Visualizing antibody affinity maturation in germinal centers. Science. 2016 Mar 4;351(6277):1048-54. doi: 10.1126/science.aad3439. Epub 2016 Feb 18.
Browne EP (2015). An Interleukin-1 beta encoding retrovirus exhibits enhanced replication in vivo. J. Virol.
Spyridon Stavrou, Daniel Crawford, Kristin Blouch, Edward P. Browne, Rahul M. Kohli and Susan R. Ross. (2014).Transgenic mice demonstrate different modes of retrovirus restriction by APOBEC3A and APOBEC3G in vivo. PLoS Pathogens.
Browne EP. (2013). TLR7 inhibits early acute retroviral infection through rapid lymphocyte responses. J Virol.
Browne EP. (2012). Regulation of B cell responses by Toll-like receptors. Immunology. Mar 24. doi: 10.1111/j.1365-2567
Nassar RA, Browne EP, Chen J, Klibanov AM (2011). Removal of HIV-1 from blood using immobilized herapin. Biotechnology letters
Browne EP (2011). TLR7 regulates the germinal center response to retroviral infection. PLoS Pathogens (October 6, 2011)
Browne EP and Littman DR (2009). Myd88 is required for an antibody response to retroviral infection. PLoS Pathog 5(2)
Browne EP, Allers C, Landau NR (2009). Inhibition of HIV-1 by APOBEC3G requires cytidine deamination. Virology
Browne EP and Littman DR (2008). Species specific restriction of Apobec3 deamination. J Virol 82(3):1305-13.
Browne EP, Li J, Chong M, Littman DR (2005). Virus-host interactions: new insights from the small RNA world. Genome Biol.;6(11):238.
Challacombe JF, Rechtsteiner A, Gottardo R, Rocha LM, Browne EP, Shenk T, Altherr MR, Brettin TS (2004). Evaluation of the host transcriptional response to human cytomegalovirus infection. Physiol Genomics. 2004 Jun 17;18(1):51-62.
Browne EP and T Shenk (2003). Human Cytomegalovirus UL83-coded pp65 virion protein inhibits anti-viral gene expression in infected cells. Proc Natl Acad Sci U S A. 2003 Sep 30;100(20):11439-44.
Browne EP, Wing B, Coleman D and T Shenk (2001) Altered Cellular mRNA Levels in Human Cytomegalovirus-Infected Fibroblasts: Viral Block to the Accumulation of Antiviral mRNAs. J Virol 75 pp12319-12330
Wing BA, Browne EP and T Shenk (2001) DNA Microarrays: A powerful new tool for the analysis of virus-host interaction. Drug Discovery Today 6 (15) suppl. pp S67-S71
Browne EP, Bellamy AR and JA Taylor (2000) Membrane-destabilizing activity of rotavirus NSP4 is mediated by a membrane-proximal amphipathic domain. J Gen Virol 84 pp1955-1959

- Edward P Browne
- Principal Investigator
- epbrowne@email.unc.edu
- Barcley T Pace
- Technician
- btpace@med.unc.edu
- Jackson J Peterson
- Graduate Student
- jjp@live.unc.edu



